1. Miller, S. L. (1953) A production of amino acids under possible primitive earth conditions. Science, 117, 528-529
2. Hutchison, C. A., Chuang, R. Y., Noskov, V. N., Assad-Garcia, N., Deerinck, T. J., Ellisman, M. H., Gill, J., Kannan, K., Karas, B. J., Ma, L., et al. (2016) Design and synthesis of a minimal bacterial genome. Science, 351, aad6253
3. Xu, C., Martin, N., Li, M., and Mann, S. (2022) Living material assembly of bacteriogenic protocells. Nature, 609, 1029-1037
4. Koshland, D. E. (2002) The seven pillars of life. Science, 295, 2215-2216
5. Smith, K. C., and Mariscal, C. (1970) Social and conceptual issues in astrobiology, Cambridge University Press
6. Cleland, C. E., and Chyba, C. F. (2007) Planets and life: The emerging science of astrobiology, Cambridge University Press
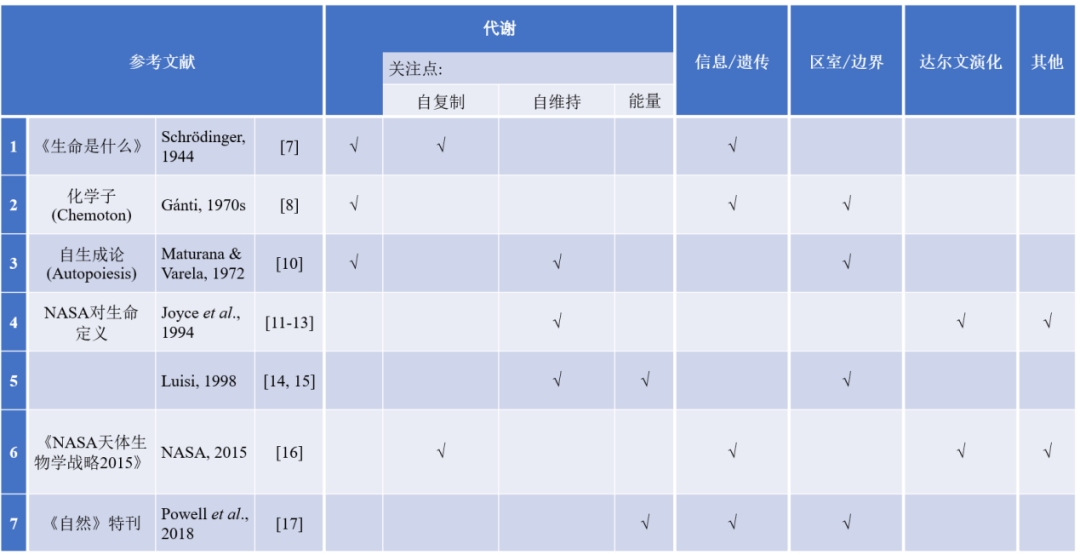
7. Schrodinger, E. (1944) What is life? And mind and matter, Cambridge University Press
8. Ganti, T. (2003) The principles of life, Oxford University Press UK
9. Shapiro, R. (2007) A simpler origin for life. Sci. Am., 296, 46–53
10. Maturana, H. R., and Varela, F. J. (1991) Autopoiesis and cognition: The realization of the living, Springer Science & Business Media
11. Joyce, C. M., and Steitz, T. A. (1994) Function and structure relationships in DNA pol ymerases. Annu. Rev. Biochem., 63, 777-822
12. Szostak, J. W., Bartel, D. P., and Luisi, P. L. (2001) Synthesizing life. Nature, 409, 387–390
13. Benner, S. A. (2010) Defining life. Astrobiology, 10, 1021-1230
14. Luisi, P. L. (1998) About various definitions of life. Biosphere, 28, 613–622
15. Damiano, L., and Luisi, P. L. (2010) Towards an autopoietic redefinition of life. Orig. Life Evol. Biosph., 40, 145-149
16. Allwood, A., Amend, J., Anbar, A., Billings, L., Blankenship, R., Boss, A., Braakman, R., Cavanaugh, C., Copley, S., Driscoll, P., et al. (2015) NASA astrobiology strategy, NASA
17. Powell, K. (2018) Biology from scratch: Built from the bottom up, synthetic cells could reveal the boundaries of life. Nature, 563, 172-175
18. Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R. Y., Algire, M. A., Benders, G. A., Montague, M. G., Ma, L., Moodie, M. M., et al. (2010) Creation of a bacterial cell controlled by a chemically synthesized genome. Science, 329, 52-56
19. Thornburg, Z. R., Bianchi, D. M., Brier, T. A., Gilbert, B. R., Earnest, T. M., Melo, M. C. R., Safronova, N., Saenz, J. P., Cook, A. T., Wise, K. S., et al. (2022) Fundamental behaviors emerge from simulations of a living minimal cell. Cell, 185, 345-360
20. Pelletier, J. F., Sun, L., Wise, K. S., Assad-Garcia, N., Karas, B. J., Deerinck, T. J., Ellisman, M. H., Mershin, A., Gershenfeld, N., Chuang, R. Y., et al. (2021) Genetic requirements for cell division in a genomically minimal cell. Cell, 184, 2430-2440
21. Shao, Y., Lu, N., Wu, Z., Cai, C., Wang, S., Zhang, L. L., Zhou, F., Xiao, S., Liu, L., Zeng, X., et al. (2018) Creating a functional single-chromosome yeast. Nature, 560, 331-335
22. Oparin, A. I. (1938) Organisms and the earth. Nature, 142, 412-413
23. Muller, H. J. (1947) The gene. Proc. R. Soc. Med., 134, 1-37
24. Lazcano, A. (2010) Which way to life? Orig. Life Evol. Biosph., 40, 161-167
25. Kauffman, S. A. (1993) The origins of order: Self-organization and selection in evolution, Oxford University Press.
26. Orgel, L. E. (2008) The implausibility of metabolic cycles on the prebiotic earth. PLoS Biol., 6, e18
27. Mizuno, T., and Weiss, A. H. (1974) Synthesis and utilization of formose sugars. Adv. Carbohydr. Chem. Biochem., 29, 173-227
28. Schwartz, A. W., and Goverde, M. (1982) Acceleration of hcn oligomerization by formaldehyde and related compounds: Implications for prebiotic syntheses J. Mol. Evol. , 18, 351-353
29. Morowitz, H. J. (1999) A theory of biochemical organization, metabolic pathways, and evolution. Complexity, 4, 39-53
30. Wächtershäuser, G. (1988) Before enzymes and templates: Theory of surface metabolism. Clin. Microbiol. Rev., 52, 452–484
31. Wächtershäuser, G. (1990) Evolution of the first metabolic cycles. Proc. Natl. Acad. Sci. USA, 87, 200-204
32. Wächtershäuser, G. (2000) Life as we don’t know it. Science, 289, 1307-1308
33. Lazcano, A., and Miller, S. L. (1999) On the origin of metabolic pathways. J. Mol. Evol., 49, 424–431
34. Delaye, L., Becerra, A., and Lazcano, A. (2005) The last common ancestor: What’s in a name? Orig. Life Evol. Biosph., 35, 537-554
35. Wolos, A., Roszak, R., Zadlo-Dobrowolska, A., Beker, W., Mikulak-Klucznik, B., Spolnik, G., Dygas, M., Szymkuc, S., and Grzybowski, B. A. (2020) Synthetic connectivity, emergence, and self-regeneration in the network of prebiotic chemistry. Science, 369, 1584
36. Troland, L. T. (1917) Biological enigmas and the theory of enzyme action. Am. Nat., 51, 321-350
37. Woese, C. R., Dugre, D. H., Saxinger, W. C., and Dugre, S. A. (1966) The molecular basis for the genetic code. Proc. Natl. Acad. Sci. USA, 55, 966–974
38. Crick, P. H. (1968) The origin of the genetic code J. Mol. Biol., 38, 367-379
39. Orgel, L. E. (1968) Evolution of the genetic apparatus. J. Mol. Biol., 38, 381-393
40. Gilbert, W. (1986) The RNA world. Nature, 319, 618
41. Copley, S. D., Smith, E., and Morowitz, H. J. (2007) The origin of the RNA world: Co-evolution of genes and metabolism. Bioorg. Chem., 35, 430-443
42. Nelson, J. W., and Breaker, R. R. (2017) The lost language of the RNA world. Sci. Signal., 10, eaam8812
43. Nissen, P., Hansen, J., Ban, N., Moore, P. B., and Steitz, T. A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920-930
44. Turk, R. M., Chumachenko, N. V., and Yarus, M. (2010) Multiple translational products from a five-nucleotide ribozyme. Proc. Natl. Acad. Sci. USA, 107, 4585-4589
45. Muller, F., Escobar, L., Xu, F., Wegrzyn, E., Nainyte, M., Amatov, T., Chan, C. Y., Pichler, A., and Carell, T. (2022) A prebiotically plausible scenario of an RNA-peptide world. Nature, 605, 279-284
46. Aravind, L., Anantharaman, V., and Koonin, E. V. (2002) Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA. Proteins, 48, 1-14
47. Danchin, A., Fang, G., and Noria, S. (2007) The extant core bacterial proteome is an archive of the origin of life. Proteomics, 7, 875-889
48. Johnston, W. K., Unrau, P. J., Lawrence, M. S., Glasner, M. E., and Bartel, D. P. (2001) RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science, 292, 1319-1325
49. Joyce, G. F. (2009) Evolution in an RNA world. Cold Spring Harb. Sym., 74, 17–23
50. Mizuuchi, R., Furubayashi, T., and Ichihashi, N. (2022) Evolutionary transition from a single RNA replicator to a multiple replicator network. Nat. Commun., 13, 1460
51. Oró, J. (1961) Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive earth conditions. Nature, 191, 1193–1194
52. Abelson, P. H. (1966) Chemical events on the primitive earth. Proc. Natl. Acad. Sci. USA, 55, 1365-1372
53. Roy, D., Najafian, K., and Schleyer, P. v. R. (2007) Chemical evolution: The mechanism of the formation
of adenine under prebiotic conditions. Proc. Natl. Acad. Sci. USA, 104, 17272–11727
54. Eschenmoser, A. (2007) On a hypothetical generational relationship between hcn and constituents of the reductive citric acid cycle. Chem. Biodivers., 4, 554-573
55. Saladino, R., Botta, G., Pino, S., Costanzo, G., and Di Mauro, E. (2012) Genetics first or metabolism first? The formamide clue. Chem. Soc. Rev., 41, 5526-5565
56. Saladino, R., Crestini, C., Pino, S., Costanzo, G., and Di Mauro, E. (2012) Formamide and the origin of life. Phys. Life Rev., 9, 84-104
57. Muchowska, K. B., Varma, S. J., and Moran, J. (2020) Nonenzymatic metabolic reactions and life’s origins. Chem. Rev., 120, 7708-7744
58. Chen, I. A., and Walde, P. (2010) From self-assembled vesicles to protocells. CSH Perspect. Biol., 2, a002170
59. Cornell, C. E., Black, R. A., Xue, M., Litz, H. E., Ramsay, A., Gordon, M., Mileant, A., Cohen, Z. R., Williams, J. A., Lee, K. K., et al. (2019) Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc. Natl. Acad. Sci. USA, 116, 17239-17244
60. Chen, I. A. (2006) The emergence of cells during the origin of life. Science, 314, 1558–1559
61. Kahana, A., and Lancet, D. (2021) Self-reproducing catalytic micelles as nanoscopic protocell precursors. Nat. Rev. Chem., 5, 870-878
62. Shapiro, R. (2007) The sudden appearance of a large self-copying molecule such as RNA was exceedingly improbable. Energy-driven networks of small molecules afford better odds as the initiators of life. Sci. Am., 296, 46–53
63. Chang, T. (2007) 50th anniversary of artificial cells: Their role in biotechnology, nanomedicine, regenerative medicine, blood substitutes, bioencapsulation, cell/stem cell therapy and nanorobotics. Artif. Cell Blood Sub. Biotech., 35, 545-554
64. Damer, B., and Deamer, D. (2015) Coupled phases and combinatorial selection in fluctuating hydrothermal pools: A scenario to guide experimental approaches to the origin of cellular life. Life, 5, 872-887
65. Mukwaya, V., Mann, S., and Dou, H. (2021) Chemical communication at the synthetic cell/living cell interface. Commun. Chem., 4, 161
66. Mukwaya, V., Zhang, P., Liu, L., Dang-i, A. Y., Li, M., Mann, S., and Dou, H. (2021) Programmable membrane-mediated attachment of synthetic virus-like nanoparticles on artificial protocells for enhanced immunogenicity. Cell Rep. Phys. Sci., 2, 100291
67. Segré, D., and Lancett, D. (2000) Composing life. EMBO Rep., 1, 217–222
68. Segré, D., Deamer, D. B.-E. D. W., and Lancet, D. (2001) The lipid world. Origins Life Evol. B., 31, 119-145
69. Serrano-Luginbühl, S., Ruiz-Mirazo, K., Ostaszewski, R., Gallou, F., and Walde, P. (2018) Soft and dispersed interface-rich aqueous systems that promote and guide chemical reactions. Nat. Rev. Chem., 2, 306-327
70. Lancet, D., Segre, D., and Kahana, A. (2019) Twenty years of “lipid world”: A fertile partnership with david deamer. Life, 9, 77
71. Kurihara, K., Tamura, M., Shohda, K., Toyota, T., Suzuki, K., and Sugawara, T. (2011) Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem., 3, 775-781
72. Neumann, J. V. (1948) The general an logical theory of automata, Papers of John Von Neumann on Computing & Computer Theory
73. Gardner, M. (1970) The fantastic combinations of john conway’s new solitaire game “life”. Sci. Am., 223, 120-123
74. Langton, C. G. (1986) Studying artificial life with cellular automata. Physica D, 22, 120-149
75. Wolfram, S. (2002) A new kind of science, Wolfram Media
76. Weihs, D., Gefen, A., and Vermolen, F. J. (2016) Review on experiment-based two- and three-dimensional models for wound healing. Interface Focus, 6, 20160038
77. Shakeel, A., and Love, P. J. (2013) When is a quantum cellular automaton (QCA) a quantum lattice gas automaton (QLGA)? J. Math. Phys., 54,
78. Cole, T., and Lusth, J. C. (2001) Quantum-dot cellular automata. Prog. Quantum. Electron., 25, 165–189
79. Langton, C. G. (1989) Artificial life, The MIT Press
80. Pilat, M. L., and Jacob, C. (2008) Creature academy: A system for virtual creature evolution. IEEE Trans. Evol. Comput., 3289-3297
81. Reynolds, C. W. (1987) Flocks, herds, and schools: A distributed behavioral model. Comput. Graph., 21, 25-34
82. Vicsek, T., Czirok, A., Ben-Jacob, E., Cohen, I. I., and Shochet, O. (1995) Novel type of phase transition in a system of self-driven particles. Phys. Rev. Lett., 75, 1226-1229
83. Hickinbotham, S., and Stepney, S. (2015) Environmental bias forces parasitism in Tierra. Proc. Eur. Conf. Artif. Life, 294-301
84. Ofria, C., and Wilke, C. O. (2004) Avida: A software platform for research in computational evolutionary biology. Artif. Life, 10, 191–229
85. Illman, S. (2001) Hilbert’s fifth problem: Review. J. Math. Sci., 105, 1843-1847
86. Holland, J. H. (1975) Adaptation in natural and artificial systems: An introductory analysis with applications to biology, control, and artificial intelligence, The MIT Press
87. Deb, K., Member, A., IEEE, Pratap, A., Agarwal, S., and Meyarivan, T. (2002) A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput., 6, 182-197
88. Szigeti, B., Gleeson, P., Vella, M., Khayrulin, S., Palyanov, A., Hokanson, J., Currie, M., Cantarelli, M., Idili, G., and Larson, S. (2014) Openworm: An open-science approach to modeling caenorhabditis elegans. Front. Comput. Neurosci., 8, 137
89. Givon, L. E., and Lazar, A. A. (2016) Neurokernel: An open source platform for emulating the fruit fly brain. PLoS One, 11, e0146581
90. Xu, C., Hu, S., and Chen, X. (2016) Artificial cells: From basic science to applications. Mater Today, 19, 516-532
91. Gomez-Marquez, J. (2021) What is life? Mol. Biol. Rep., 48, 6223-6230
92. Liu, Y., and Sumpter, D. J. T. (2018) Mathematical modeling reveals spontaneous emergence of self-replication in chemical reaction systems. J. Biol. Chem., 293, 18854-18863
93. Stewart, J. E. (2018) The origins of life: The managed-metabolism hypothesis. Found. Sci., 24, 171-195
94. Ameta, S., Matsubara, Y. J., Chakraborty, N., Krishna, S., and Thutupalli, S. (2021) Self-reproduction and darwinian evolution in autocatalytic chemical reaction systems. Life, 11, 308
95. Kauffman, S. (1995) At home in the universe: The search for laws of self-organization and complexity, Oxford University Press
96. Hordijk, W., and Steel, M. (2012) Autocatalytic sets extended: Dynamics, inhibition, and a generalization. J. Syst. Chem., 3, 5
97. Liu, Y., Hjerpe, D., and Lundh, T. (2020) Side reactions do not completely disrupt linear self-replicating chemical reaction systems. Artif. Life, 26, 327-337
98. Liu, Y. (2020) On the definition of a self-sustaining chemical reaction system and its role in heredity. Biol. Direct., 15, 15
99. Fontana, W., and Buss, L. W. (1994) “The arrival of the fittest”: Toward a theory of biological organization. Bull. Math. Biol., 56, 1–64
100. Dittrich, P., and di Fenizio, P. S. (2007) Chemical organisation theory. Bull. Math. Biol., 69, 1199-1231
101. Hordijk, W., Steel, M., and Dittrich, P. (2018) Autocatalytic sets and chemical organizations: Modeling self-sustaining reaction networks at the origin of life. New J. Phys., 20, 015011
102. Sousa, F. L., Hordijk, W., Steel, M., and Martin, W. F. (2015) Autocatalytic sets in E. coli metabolism. J. Syst. Chem., 6, 4
103. Semenov, S. N., Kraft, L. J., Ainla, A., Zhao, M., Baghbanzadeh, M., Campbell, V. E., Kang, K., Fox, J. M., and Whitesides, G. M. (2016) Autocatalytic, bistable, oscillatory networks of biologically relevant organic reactions. Nature, 537, 656-660
104. Sadownik, J. W., Mattia, E., Nowak, P., and Otto, S. (2016) Diversification of self-replicating molecules. Nat. Chem., 8, 264-269
105. Monreal Santiago, G., Liu, K., Browne, W. R., and Otto, S. (2020) Emergence of light-driven protometabolism on recruitment of a photocatalytic cofactor by a self-replicator. Nat. Chem., 12, 603-607
106. Nanda, J., Rubinov, B., Ivnitski, D., Mukherjee, R., Shtelman, E., Motro, Y., Miller, Y., Wagner, N., Cohen-Luria, R., and Ashkenasy, G. (2017) Emergence of native peptide sequences in prebiotic replication networks. Nat. Commun., 8, 434
107. Peng, Z., Linderoth, J., and Baum, D. A. (2022) The hierarchical organization of autocatalytic reaction networks and its relevance to the origin of life. PLoS Comput. Biol., 18, e1010498
108. Xavier, J. C., Hordijk, W., Kauffman, S., Steel, M., and Martin, W. F. (2020) Autocatalytic chemical networks at the origin of metabolism. Proc. Biol. Sci., 287, 20192377
109. Adam, Z. R., Fahrenbach, A. C., Jacobson, S. M., Kacar, B., and Zubarev, D. Y. (2021) Radiolysis generates a complex organosynthetic chemical network. Sci. Rep., 11, 1743
110. Keller, M. A., Turchyn, A. V., and Ralser, M. (2014) Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible archean ocean. Mol. Syst. Biol., 10, 725
111. Pedersen, R. B., Rapp, H. T., Thorseth, I. H., Lilley, M. D., Barriga, F. J., Baumberger, T., Flesland, K., Fonseca, R., Fruh-Green, G. L., and Jorgensen, S. L. (2010) Discovery of a black smoker vent field and vent fauna at the arctic mid-ocean ridge. Nat. Commun., 1, 126
112. Georgieva, M. N., Little, C. T. S., Maslennikov, V. V., Glover, A. G., Ayupova, N. R., and Herrington, R. J. (2021) The history of life at hydrothermal vents. Earth-Sci. Rev., 217, 103602
113. Martin, W., and Russell, M. J. (2003) On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B Biol. Sci., 358, 59-83
114. Martin, W., and Russell, M. J. (2007) On the origin of biochemistry at an alkaline hydrothermal vent. Philos. Trans. R. Soc. Lond. B Biol. Sci., 362, 1887-1925
115. Roldan, A., Hollingsworth, N., Roffey, A., Islam, H. U., Goodall, J. B., Catlow, C. R., Darr, J. A., Bras, W., Sankar, G., Holt, K. B., et al. (2015) Bio-inspired CO2 conversion by iron sulfide catalysts under sustainable conditions. Chem. Commun., 51, 7501-7504
116. Camprubi, E., Jordan, S. F., Vasiliadou, R., and Lane, N. (2017) Iron catalysis at the origin of life. IUBMB Life, 69, 373-381
117. Belthle, K. S., and Tüysüz, H. (2022) Linking catalysis in biochemical and geochemical CO2 fixation at the emergence of life. ChemCatChem, e202201462
118. Varma, S. J., Muchowska, K. B., Chatelain, P., and Moran, J. (2018) Native iron reduces CO2 to intermediates and end-products of the acetyl-CoA pathway. Nat. Ecol. Evol., 2, 1019-1024
119. Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., and Martin, W. F. (2016) The physiology and habitat of the last universal common ancestor. Nat. Microbiol., 1, 16116
120. Kelley, D. S., Karson, J. A., Fruh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., Hayes, J. M., Schrenk, M. O., Olson, E. J., Proskurowski, G., et al. (2005) A serpentinite-hosted ecosystem: The lost city hydrothermal field. Science, 307, 1428-1434
121. Kitadai, N., and Maruyama, S. (2018) Origins of building blocks of life: A review. Geosci. Front., 9, 1117-1153
122. Koonin, E. V., and Martin, W. (2005) On the origin of genomes and cells within inorganic compartments. Trends Genet., 21, 647-654
123. Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D. V., Galperin, M. Y., and Koonin, E. V. (2012) Origin of first cells at terrestrial, anoxic geothermal fields. Proc. Natl. Acad. Sci. USA, 109, E821-E830
124. Patel, B. H., Percivalle, C., Ritson, D. J., Duffy, C. D., and Sutherland, J. D. (2015) Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem., 7, 301-307
125. Deamer, D., Damer, B., and Kompanichenko, V. (2019) Hydrothermal chemistry and the origin of cellular life. Astrobiology, 19, 1523-1537
126. Cairns-Smith, A. G. (1987) Genetic takeover: And the mineral origins of life, Cambridge University Press
127. Zimmer, C. (2004) What came before DNA? Discover.
128. Huang, W., and Ferris, J. P. (2006) One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc., 128, 8914-8919
129. Kloprogge, J. T. T., and Hartman, H. (2022) Clays and the origin of life: The experiments. Life, 12, 259
130. Baum, D. A. (2018) The origin and early evolution of life in chemical composition space. J. Theor. Biol., 456, 295-304
131. Vincent, L., Berg, M., Krismer, M., Saghafi, S. S., Cosby, J., Sankari, T., Vetsigian, K., Ii, H. J. C., and Baum, D. A. (2019) Chemical ecosystem selection on mineral surfaces reveals long-term dynamics consistent with the spontaneous emergence of mutual catalysis. Life, 9, 80
132. Xu, Z., Hueckel, T., Irvine, W. T. M., and Sacanna, S. (2021) Transmembrane transport in inorganic colloidal cell-mimics. Nature, 597, 220-224
133. Thoren, H., and Gerlee, P. (2010) Weak emergence and complexity, MIT Press
134. Kolchinsky, A., and Wolpert, D. H. (2018) Semantic information, autonomous agency and non-equilibrium statistical physics. Interface Focus, 8, 20180041
135. Liu, Y., Mathis, C., Bajczyk, M. D., Marshall, S. M., Wilbraham, L., and Cronin, L. (2021) Exploring and mapping chemical space with molecular assembly trees. Sci. Adv., 7, eabj2465
136. Liu, Y., Di, Z., and Gerlee, P. (2022) Ladderpath approach: How tinkering and reuse increase complexity and information. Entropy, 24, 1082
137. Baum, D. A., and Lehman, N. (2017) Life’s late digital revolution and why it matters for the study of the origins of life. Life, 7, 34
138. Szathmary, E. (2000) The evolution of replicators. Philos. Trans. R. Soc. Lond. B Biol. Sci., 355, 1669-1676
139. Powell, K. (2018) Biology from scratch built: From the bottom up, synthetic cells could reveal the boundaries of life. Nature, 563, 172-175
140. Deshpande, S., Caspi, Y., Meijering, A. E., and Dekker, C. (2016) Octanol-assisted liposome assembly on chip. Nat. Commun., 7, 10447
141. Weiss, M., Frohnmayer, J. P., Benk, L. T., Haller, B., Janiesch, J. W., Heitkamp, T., Borsch, M., Lira, R. B., Dimova, R., Lipowsky, R., et al. (2018) Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater., 17, 89-96
142. Steel, M., Xavier, J. C., and Huson, D. H. (2020) The structure of autocatalytic networks, with application to early biochemistry. J. R. Soc. Interface, 17, 20200488
143. Lin, G.-M., Warden-Rothman, R., and Voigt, C. A. (2019) Retrosynthetic design of metabolic pathways to chemicals not found in nature. Curr. Opin.Sys. Biol., 14, 82-107
144. Mikulak-Klucznik, B., Golebiowska, P., Bayly, A. A., Popik, O., Klucznik, T., Szymkuc, S., Gajewska, E. P., Dittwald, P., Staszewska-Krajewska, O., Beker, W., et al. (2020) Computational planning of the synthesis of complex natural products. Nature, 588, 83-88
145. Burger, B., Maffettone, P. M., Gusev, V. V., Aitchison, C. M., Bai, Y., Wang, X., Li, X., Alston, B. M., Li, B., Clowes, R., et al. (2020) A mobile robotic chemist. Nature, 583, 237-241
146. Steiner, S., Wolf, J., Glatzel, S., Andreou, A., Granda, J. M., Keenan, G., Hinkley, T., Aragon-Camarasa, G., Kitson, P. J., Angelone, D., et al. (2019) Organic synthesis in a modular robotic system driven by a chemical programming language. Science, 363, 144
147. Wu, Z., Kan, S. B. J., Lewis, R. D., Wittmann, B. J., and Arnold, F. H. (2019) Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl. Acad. Sci. USA, 116, 8852-8858
148. Yang, K. K., Wu, Z., and Arnold, F. H. (2019) Machine-learning-guided directed evolution for protein engineering. Nat. Methods, 16, 687-694